Precision Medicine of Brain Cancer
a Weakly-Supervised Transfer Learning model for personalized predictive modeling of tumor cell density
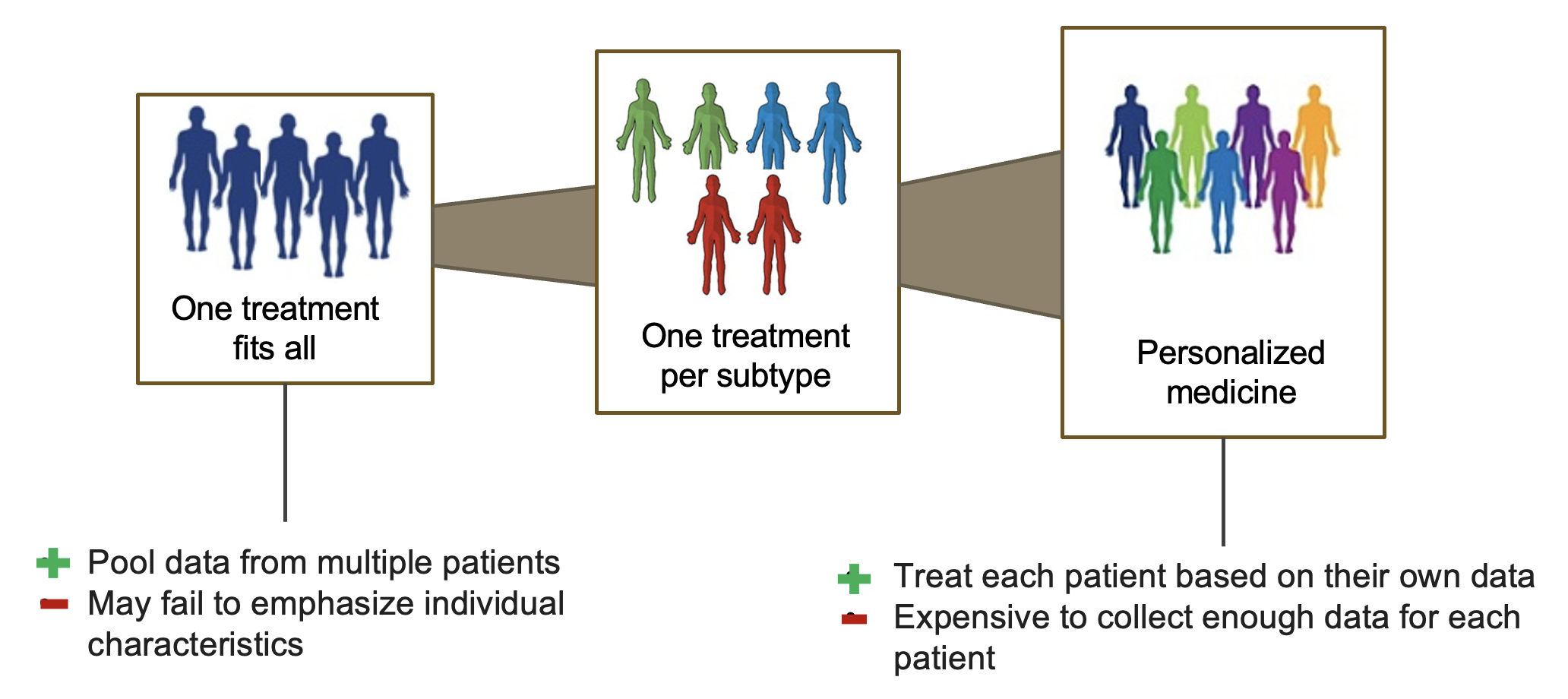
Precision medicine aims to provide diagnosis and treatment accounting for individual differences. To develop machine learning models in support of precision medicine, personalized or patient-specific models are expected to have better performance than one-model-fits-all approaches.
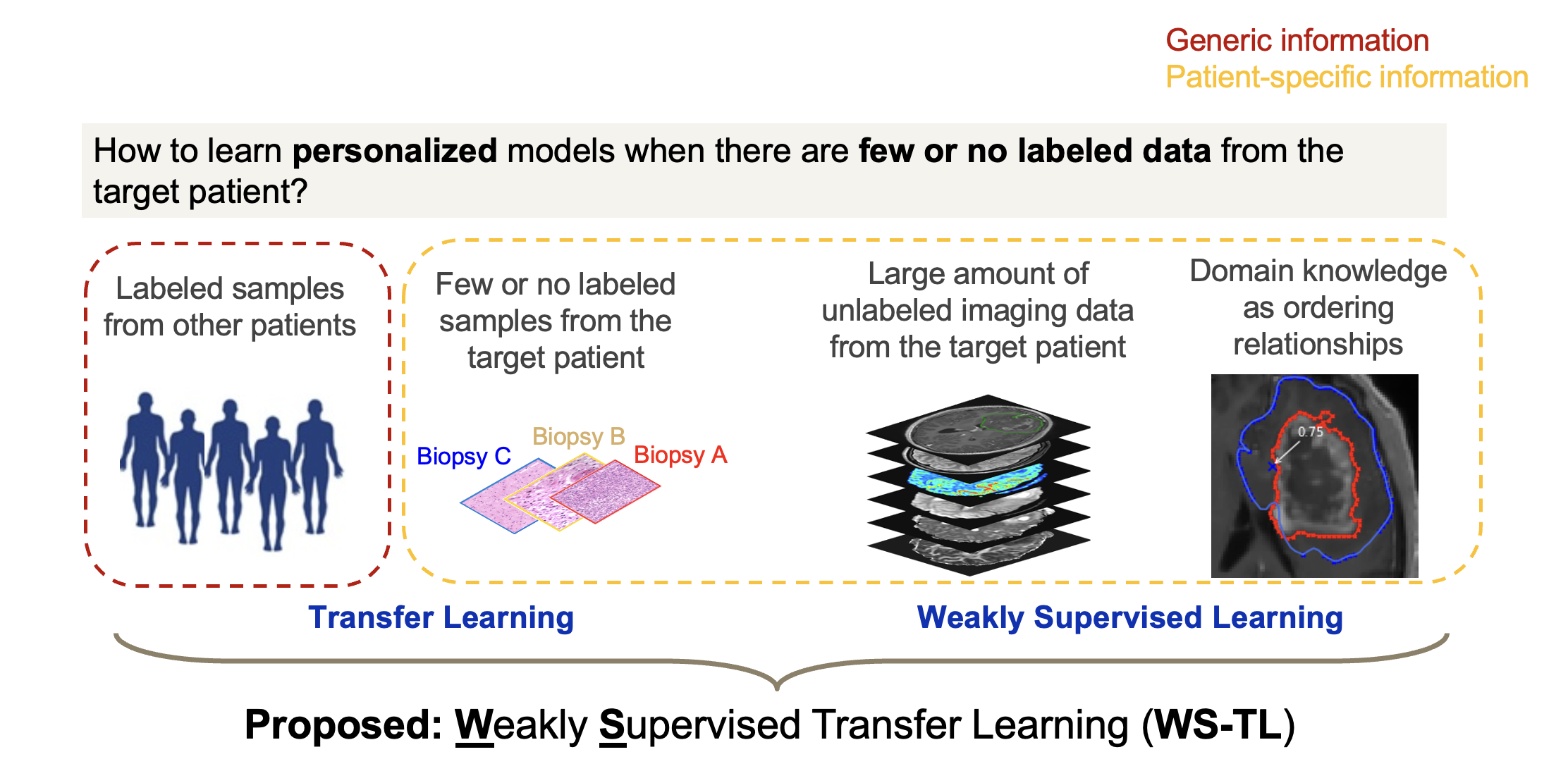
Example Application: Glioblastoma (GBM) is the most aggressive type of brain cancer with a median survival of only 15 months. One challenge of treatment is the intratumoral heterogeneity -tumor is a mosaic, with subpopulations of cells with different genetic alterations and thus different biological behaviors. It is important to know the regional Tumor Cell Density (TCD) —the percentage of tumor cells within a regional tissue sample— so that treatment can be optimized, i.e., regions with higher TCD can be treated more aggressively to prevent tumor growth whereas regions with lower TCD can be treated less aggressively to avoid over-damaging of the brain. To know the TCD of a specific region, the gold-standard approach is to acquire a biopsy sample from that region and obtain TCD measurement through histopathologic analysis. However, due to the invasive nature of biopsy, only a few biopsy samples can be acquired from each patient, leaving many regions where TCD remains unknown. To tackle this challenge, a machine learning model can be trained to predict regional TCD based on imaging. Since imaging is non-invasive and can portray the whole brain, using the trained machine learning model allows for generating a predictive TCD map for each patient to guide individualized treatment.
A significant challenge to build personalized prediction models, however, is the limited number of labeled samples for each individual due to cost, availability, and other practical constraints. Transfer Learning (TL) addresses this challenge by utilizing data from other patients with the same disease (i.e., the source domain) to create a model tailored to the individual (i.e., the target domain). However, existing TL algorithms require a significant number of labeled target samples to effectively adapt the source model, limiting TL’s effectiveness in cases where target domain data is extremely sparse, such as in predicting spatial biomarkers for brain cancer.
We developed a new Weakly Supervised Transfer Learning (WSTL) model that enables building personalized model even with very few or no labeled samples from the target patient. WS-TL leverages domain knowledge to generate weakly labeled samples from unlabeled data, creating patient-specific information that complements generalized data from other patients. This fusion of labeled source-domain data (e.g. from other patients of the same disease) with knowledge-informed, weakly labeled target-domain data (e.g. from the patient to be predicted) results in robust personalized predictive models. Theoretical analysis shows that WSTL has beneficial theoretical properties such as solution sparseness and robustness to outliers.
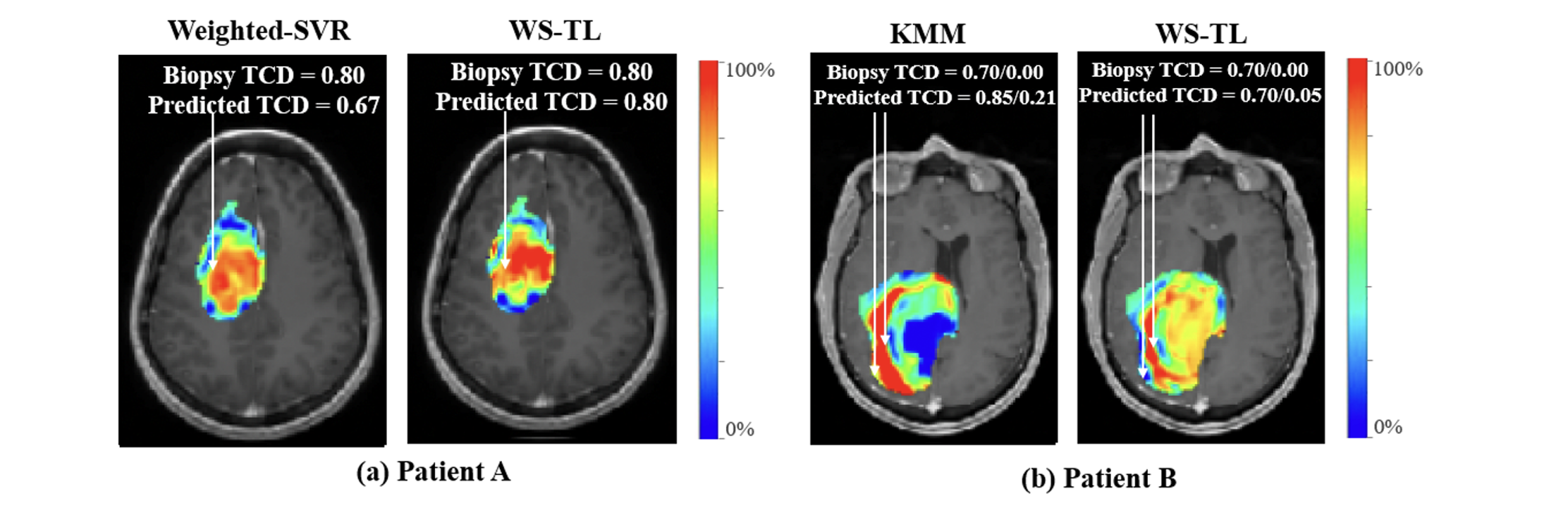
Applying WSTL to predict regional Tumor Cell Density (TCD) from Magnetic Resonance Imaging of Glioblastoma patients, we achieve significantly higher accuracy to other one-fits-all and personalized modeling methods, showcasing WSTL’s potential for guiding precision treatments in brain cancer via non-invasive neuroimaging.
Publications:
- Mao L, Wang H, Li J. Knowledge-informed machine learning for cancer prognosis and predictions: a review. (major revision at IEEE T-ASE). (website) (paper)
- Mao L, Wang L, Hu L, Eschbacher J, Leon GD, Singleton K, Curtin W, Urcuyo A, Sereduk J, Tran L, Hawkins A, Swanson K, Li J. Weakly supervised transfer learning with application in precision medicine. IEEE Transactions on Automation Science and Engineering. doi:10.1109/TASE.2023.3323773 (paper) (🏆 Best Student Paper, IISE DAIS 2022)
- Lewis, EM, Mao, L, Wang, L, Swanson, KR, Barajas, RF, Li, J, … & Plaisier, CL (2023). Revealing the biology behind MRI signatures in high grade glioma. medRxiv.